Welcome to Drop-seq!
Drop-seq is a technology we developed to enable biologists to analyze RNA expression genome-wide in thousands of individual cells at once. We first described this in a 2015 paper in Cell. Though commercial implementations of droplet-based single-cell RNA-seq also now exist, we have made Drop-seq open-source and want to make sure that any lab can build their own system. The materials for constructing a Drop-seq setup in one’s own lab can be obtained for about $6,000. The reagents for performing Drop-seq cost about 6 cents per cell.
This site provides instructions, videos and software with which any scientist can build a Drop-seq setup, do Drop-seq experiments, and prepare the resulting data for a variety of downstream analyses. Hundreds of labs have now built their own Drop-seq systems.
We also have a Google Groups forum dedicated to discussing Drop-seq implementations and experiments. More than 800 scientists currently subscribe. To participate, please send an email to dropseq@gmail.com and we’ll put you on the list.
We hope you do amazing things with Drop-seq!
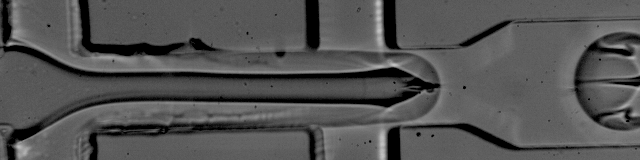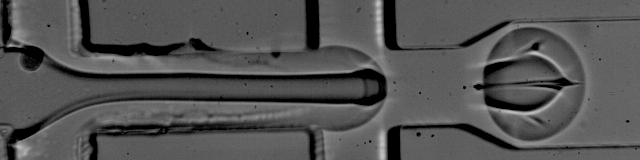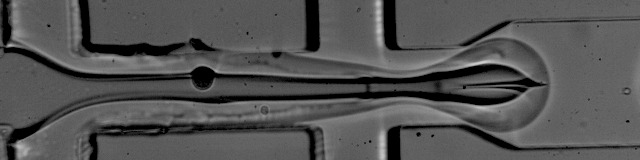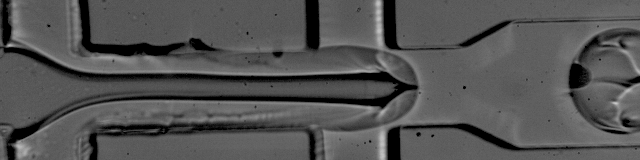
Image courtesy of Patrick Stumpf, Matthew Rose-Zerilli, Rosanna Smith, Martin Fischlechner & Jonathan West at the Centre for Hybrid Biodevices & Cancer Sciences Unit at the University of Southampton
Laboratory resources

A Drop-seq setup can be built from widely available laboratory equipment costing less than ten thousand dollars. We used this one setup to perform all of the experiments in the Cell paper.
Here is our latest experimental protocol, which includes instructions for building a Drop-seq setup:
Online-Dropseq-Protocol-v.-3.1-Dec-2015.pdf (49978 downloads)
This protocol reflects our current, optimized Drop-seq protocol. (It does not precisely match the methods section of our Cell paper, because we continue to improve the protocols and place improvements online.) This protocol also includes hints, suggestions, and images.
We want this web site to be a “living protocol”. As people begin to adopt Drop-seq and we learn about their experiences, we will add additional hints and tips to the protocol, releasing updated protocols with a new date. We will increment the version number only when we change something substantive about the recommendation for a protocol step. Please contact us with questions or suggestions – we look forward to hearing about new optimizations!
Materials
- Barcoded beads. In order to have a scalable, commercial solution available for users, we taught scientists at a local company (Chemgenes) the split-pool oligo synthesis process we describe in the Cell paper. We have extensively tested these beads in Drop-seq. We have asked Chemgenes to make these beads available directly to anyone who orders them. The beads have catalog number CSO-2011.
- Microfluidic devices
- Here you can download the CAD file for the co-flow microfluidics device, which was designed by our colleague Anindita Basu in the labs of Aviv Regev and David Weitz. This is a passive-flow PDMS device that can be synthesized in any academic or commercial microfluidics facility.
- Alternatively, microfluidic devices can be purchased ready-made from two companies, Nanoshift and FlowJEM. Simply tell them you’re interested in ordering the latest Drop-seq devices for a quote. See our FAQ page for more information.
Videos and FAQs
These tutorials, images and diagrams may be helpful in building your own Drop-seq setup and doing Drop-seq experiments in your lab.
We have also created a YouTube channel with a variety of videos to help scientists through the steps that most benefit from watching.
This FAQ provides also provides supplementary information.
Supplemental protocols
Here are two supplemental protocols you might find helpful:
• Measuring-Droplet-Volume-in-Home-Made-Devices.pdf (12197 downloads)
• Aquapel-Treatment-of-Drop-seq-Devices.pdf (12796 downloads)
Software
Our latest software, and cookbook for core computational analysis of Drop-seq data, which shows you how to transform raw sequence data into an expression measurement for each gene in each individual cell is now open-source on GitHub. The software was developed by Jim Nemesh and Alec Wysoker in our lab. Releases can be found here.
We add updates (and revise the protocol date) whenever we see opportunities to make something better, faster or more clear. We will increment the version number whenever we revise the software or change something substantive about the recommended series of steps.
We further analyze the resulting data using our colleague Rahul Satija‘s fantastic single-cell analysis package Seurat, which produced all of the clustering analyses in the Cell paper.
Data resources
Here are alignment data resouces (the same ones as described in our paper) for MOUSE, HUMAN, and MIXED (human+mouse). Scientists in the Drop-seq community have also now created such resources for many other organisms.
Example data sets
We have deposited all of the raw and processed data sets from the Drop-seq experiments described in the Cell paper (Macosko et al, 2015) in Gene Expression Omnibus, accession GSE63473. Cluster assignments for the 44,808 retinal cells in the paper are here.
Questions
Please email questions to dropseq@gmail.com. We’ll do our best to answer them!
Melissa Goldman, Jim Nemesh, Evan Macosko, Alec Wysoker, Steve McCarroll