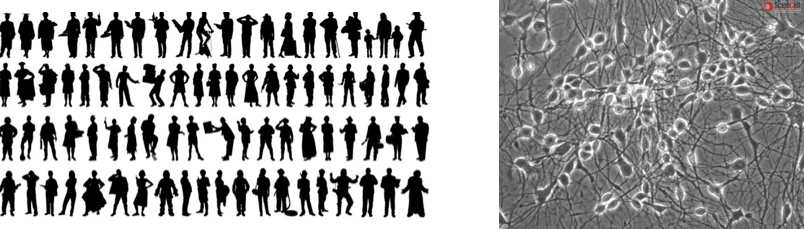
Massively mosaic experimental systems
Mosaic Experimental Systems are new experimental systems that enable population-scale analyses of how genetic variation shapes cellular phenotypes. These systems have been developed collaboratively by scientists in the labs of Steve McCarroll and Kevin Eggan.
In Mosaic Experimental Systems, we co-culture cells from many different people in a single well, dish, or organoid. These genetically diverse cells form networks, tissues, and other kinds of cell populations. We analyze these systems using Drop-seq, and use the transcribed sequence polymorphisms (SNPs) to infer which cell came from which donor.
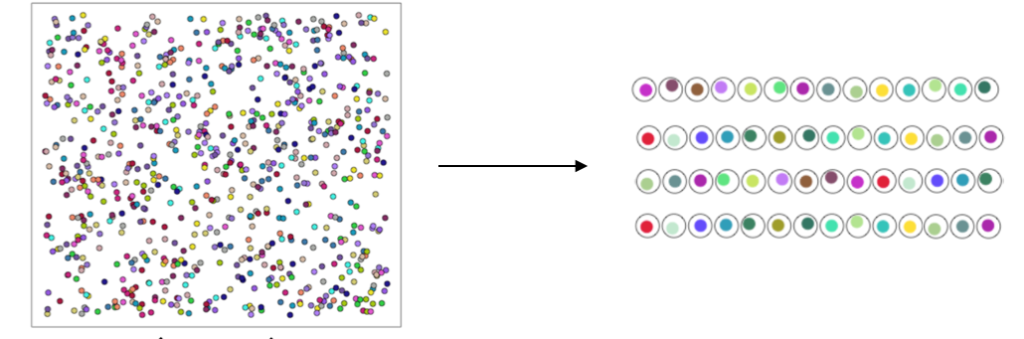
Population-scale experiments allow us to appreciate how genetic variation and other donor-specific factors give rise to variation in cellular phenotypes. The Mosaic Experimental Systems allow us to perform population-scale experiments in a single reaction – in a manner that is efficient, rigorous, and internally controlled.
Our labs have collaborated closely to develop these systems. We have a weekly lab meeting together, at which we plan new experiments and discuss ongoing analyses.
Problems we’ve solved along the way
Computational methods. An initial question was whether we could definitively identify the donor-of-origin of individual cells from a large, genetically diverse pool of hundreds of candidate donors. Jim Nemesh developed computational methods for doing this accurately, and tested these methods on both in silico and in vitro cell mixtures.
Experimental methods. Jana Mitchell developed ways to grow scores of cell lines together while maintaining diversity and representation. Along the way, she solved problems around variation in cells’ growth rates, often caused by mutations the cells acquire in culture (we recently described some of these). We increasingly do experiments in stable, genetically diverse pools.
Support
Throughout 2015-2017, our work developing Mosaic Experimental Systems has been supported by the Broad Institute’s Stanley Center for Psychiatric Research.
In September 2017, we received an NIMH Convergent Neuroscience award to scale up this research and address a range of neurobiological questions. The grant will provide more than $12 million of funding over the coming five years. The Stanley Center is also supporting the scale-up of the project.
Project expansion
We are expanding these analyses to cells from 1,000 donors. To do this, we have formed a partnership with the California Institute for Regenerative Medicine (CIRM), which has derived iPSCs from 3,000 different donors. The lines are currently being expanded and whole genome sequenced for the project.
Scientific resources for the community
Our goal is to create entirely new play books for experimental biology – and to share them widely, in the hopes of making new kinds of science possible. We plan to make computational software and experimental protocols available on this web site soon. We will also broadly share the whole genome sequences of the CIRM lines, which are available to all researchers.
The team


Getting involved
We welcome contributions from scientists at any point in their training – from postdocs to students to staff scientists There are many ways to contribute:
• Experimental biology. We hope to use Mosaic Experimental Systems to address diverse questions in neuroscience – from protein localization to anatomy, physiology, and connectivity. There are many new opportunities to design and execute experiments that exploit the power of these systems to reveal how genetic variation shapes the biology of neurons, glia, and other cell types.
• Computational and statistical analysis. Mosaic Experimental Systems are powered by computation and data science. We are always looking for ways to (i) further expand the capability and power of these systems, (ii) apply these systems to new kinds of biological problems, and (iii) analyze the data from these systems in new and creative ways.
If you are interested in getting involved, please send a letter and CV to Steve and Kevin at MosaicExperiments@gmail.com.